 Author
Author  Correspondence author
Correspondence author
Molecular Soil Biology, 2016, Vol. 7, No. 10 doi: 10.5376/msb.2016.07.0010
Received: 25 Mar., 2016 Accepted: 12 Apr., 2016 Published: 12 Apr., 2016
Sun Y.J., Ye X.X., Liu P.P. and Lee I., 2016, Analysis of Sequence from Chloris virgata for Function Classification, Molecular Soil Biology, 7(9): 1-8 (doi: 10.5376/msb.2016.07.0010)
Chloris virgata Swartz (C. virgata) is an one-year-old herbaceous plant of Gramineae. It has high adaptability and can survive in highly saline soils. We randomly selected 3168 clones from a C. virgata cDNA library from NaHCO3-stressed. By finding Restriction Enzyme cutting site to remove vectors of the sequences and delete the sequences without Restriction Enzyme cutting site, we obtained 3043 clones finally. Gene Ontology (GO) Slim annotations were obtained by BLAST2GO and it was found that 8 genes of them were annotated with GO terms “response to stress” indicating that these genes were likely to function in tolerance mechanism of C. virgata, Setaria italica was the highest homology to sequences of C. virgata. Clusters of Orthologous Group(COG) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis showed that, most of these genes involved in “cellular process”, “General function prediction only” and “ Ribosome”.
Introduction
According to the second national soil survey, Salinization of agricultural acreage in an area of 9.209 million hm2, Mainly in the northwest, north, northeast and coastal areas in china. The saline-alkali soils of northeast China contain high amounts of NaHCO3 and Na2CO3, The pH of the soil is more than 9 because of hydrolysis of these salts (Wang Jiali et al., 2011; Chao DongHui, 2012)
Soil salinization is a severe problem encountered worldwide in agricultural production that causes abiotic stress to plants and lowers crop growth and yields, thus impacting food security (Asish Kumar Parida, 2004). It also affects forestry and animal husbandry and even the whole environmental quality,thus inhibiting human living and development (Dai Gaoxing et al., 2003; Dong Lili et al., 2015). Plant can produce a variety of different mechanisms to adapt to the salt stress environment, such as cell automatic adjustment, stress damage control and repair, detoxification and growth regulators (Zhu, 2001). Thus, elucidating the molecular basis of salt stress signal transduction pathways and salt tolerance mechanisms is fundamental to understanding the biology of salt-tolerant plants. In order to support strategies for the design, genetic engineering and breeding of salt tolerant crops (Qingmei Guan et al., 2013).
C. virgateis is one of the wild plants that can survive in the saline-alkali soil areas of northeast China It is a high seed yield, and a quick germinate in summer rainy period, and it forms a single dominant species easily. Therefore C. virgata is a pioneer plant to improve the abandoned to alkaline land and alkaline grasslands, occupying an important position in the sand and aquatic, abandoned land, halophytes series succession.
Currently, there are many reports about Salt aspects pasture or crop quality,but rarely research on the role of salinity for C. virgata. Few documents that are related to C. virgata were found. Previous study suggested that the root adaptation to PH is one of the important factors for alkali stress tolerance in C. virgata (Changyou Li et al., 2009). A metallothionein-1 protein played as a function in tolerating oxidative, salinity and carbonate stress (Nishiuchi S et al., 2007). Sources of salts stresses were from NaCl (Xi Zhang et al., 2011), NaHCO3 (Shunsaku Nishiuchi et al., 2010) and Na2CO3 (Jin et al., 2008). However, the saline-alkali tolerance mechanisms is still unknown for C. virgata.
In this work to better understand the genes that confer saline-alkali stress tolerance of C. virgata, we randomly selected 3168 clones in a cDNA library from NaHCO3-stressed C. virgata for functional classification. Our results showed that the genes induced by the NaHCO3 stress were highly homologous to the gene sequences from Setaria italica. Most of these genes involved in “cellular process”, “General function prediction only” and “Ribosome”.
Materials and Methods
We selected 3168 highly expression cloned sequences in a cDNA library from NaHCO3-stressed C. virgata. By adding the upstream primer and downstream primer, we determine the restriction enzyme cutting site to remove vectors of the sequences and delete the sequences without restriction enzyme cutting site. We obtained 3043 clones finally.
GO Slim function classification
All of the unigenes were compared with sequences in the Nr database, using BLASTX with a cutoff e-value of 1E−3, and the GO Slim annotation of the C. virgata. unigenes was performed with BLAST2GO (Conesa A et al., 2005).
Gene Ontology (GO) (http://www.geneontology.org/) is used to standardize representation of genes across species and provides a set of structured and controlled vocabularies for annotating genes, gene products, and sequences (Consortium, 2008).
COG function classification
COG used for a database built on phylogenetic relationships of protein sequences from 66 genomes, including bacteria, plants and animals. Individual proteins or paralogs from at least three lineages are categorized in each COG to represent an ancient conserved domain (Yiou Pan et al., 2015).
Pathway function classification
The Kyoto encyclopedia of genes and genomes (KEGG) analysis using KEGG Automatic Annotation Server (KAAS) (http://www.genome.jp/tools/kaas/) combined with domain analysis of the assembled transcripts for revealing putative members of secondary metabolism pathways.
Results and Discussion
Annotation of predicted proteins with NR database
All of the unigenes were compared with sequences in the Nr database, using BLASTX with a cutoff e-value of E−3. 2,731 unigenes (89.7%) of all unigene sequences could analyze and obtained Blastx result (Fig. 1).
 Figure 1 The statistics of available data obtained from cDNA library from NaHCO3-stressed C. virgate in the NR database |
The E-value distribution of the top hits in the NR database showed that 41.62% of the mapped sequences had strong homology with the E-value <1.0E−90, whereas 58.38% of the homolog sequences ranged from 1.0E−4 to 1.0E−90 (Fig.2A). The distribution of similarity values showed that 83.42 % of the query sequences had a similarity of more than 80 %, while 16.59% of the hits had a similarity ranging from 44 % to 80 % (Fig.2B). In terms of species distribution, the majority of the annotated sequences corresponded to the known nucleotide sequences of plant species, 34.64 % of the sequences showed the highest homology to sequences from Setaria italica, followed 19.57% from by Zea mays, 12.85 % from Sorghum bicolor, 8.25% from Oryza sativa with and 3.45% from Oryza brachyantha (Fig.2C). The three species of the most BLAST hits belonged to the grass family, indicating that the sequences of the C. virgata transcripts obtained in the present study were annotated properly.
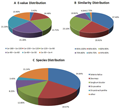 Figure 2 Characteristics of homology search for Illumina sequences against the NR database. A) E-value distribution of BLAST hits for matched unique sequences with an E-value cut-off of 1.0E−3. B) Similarity distribution of the top BLAST hits for each unique sequence. C) Distribution of homologous Species |
Functional annotation and classification of the sequences
Because Oryza sativa's (OS) comments more complete information at this stage, so we downloaded the protein sequences of OS from the NCBI, in order to build a rice protein database for blastx comparison. All sequences of the unigenes were compared with the OS database, using BLASTX with a cutoff e-value of E−3, 2,570 unigenes (84.5%) of all unigene sequences could analyze and obtained Blastx result (Fig. 3).
 Figure 3 The statistics of available data obtained from cDNA library from NaHCO3-stressed C. virgate in the OS database |
Then GO Slim annotation of the C. virgata unigenes was performed with BLAST2GO (Conesa A et al., 2005) (Fig 1).GO assignments can provide standardized vocabulary for assigning functions of the uncharacterized sequences and hence were used to classify the functions of the predicted genes (Harris MA et al., 2004) . In total, 957 sequences were categorized into 45 level-2 functional groups, including biological processes, cellular components, and molecular functions (Fig. 4).
 Figure 4 Gene Ontology classification. as three main categories: cellular component ,molecular function and biological process |
The dominant subcategories were “cell” (781) among cellular components, “binding” (423) among molecular function, and “cellular process” (722) among biological processes. There were that 432 genes that were annotated with GO Slim annotation “response to stress”, indicating these genes that were likely to function in tolerance mechanism of C. virgata. Then, the 432 genes were analyzed with David (https://david.ncifcrf.gov/), we obtained functional Annotation related to salt stress futher. (Table 1), there were 5 genes that were annotated with “stress response” (Table 2).
 Table 1 Functional annotation with DAVID |
 Table 2 Genes related to "stress response" with DAVID |
COG classification was used to further evaluate the effectiveness of the annotation process and the completeness of the transcriptome library. A COG database was built from classifications of phylogenetic relationships, consisting of protein sequences encoded in 21 complete genomes including those of bacteria, eukaryotes and algae (Dutkowski JJ, 2007) Each COG classification consists of groups of paralogs or individual proteins from at least three lineages, and thus corresponds to an ancient conserved domain (Cheng-Ying Shi et al., 2011). Sequences were assigned to the COG classification. As some of these sequences were annotated with multiple COG functions, altogether 1,588 functional annotations were generated. Among the 23 COG categories, the cluster for “General function prediction only” (264, 16.62 %) represented the largest group, followed by “Translation, ribosomal structure and biogenesis” (234, 14.74 %), “Posttranslational modification, protein turnover, chaperones” (149, 9.38 %) and “Carbohydrate transport and metabolism” (133, 8.38 %). The categories of “Chromatin structure and dynamics” and “Intracellular trafficking, secretion, and vesicular transport” were the smallest groups (Fig. 5).
 Figure 5 COG functional classification of all unigenes sequences. 1,047(34.41 %) unigenes showed significant similarity to sequences in the COG databases and were clustered into 23 categories |
The KEGG pathway database can facilitate a systematic understanding of the molecular interactions among genes, in terms of networks (Du J et al., 2014). To investigate the involvement of selected genes in metabolism pathway, a search was performed using KEGG Automatic Annotation Server (KASS, http://www.genome.jp/tools/kaas/). KAAS provides functional annotation of genes by BLAST or GHOST comparisons against the manually curated KEGG GENES database. All the sequences were annotated into Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways with KAAS (Moriya Y et al., 2007), using the Single-directional Best Hit (SBH) method. The result contains KO (KEGG Orthology) assignments and automatically generated KEGG pathways against KEGG database (Lin CMW, 2012).
Annotated sequences were matched in the KEGG database and were assigned to 271 active pathways. The five largest pathway groups were Ribosome [ko03010, 80 (6.90 %)], Carbon metabolism [ko01200, 45 (3.88 %)], Biosynthesis of amino acids [ko01230, 41(3.53 %)], Oxidative phosphorylation [ko00190, 33 (2.84 %)], Photosynthesis [ko00195, 23 (1.98%)] and Protein processing in endoplasmic reticulum [ko04141, 23(1.98%)] (Fig. 6).
 Figure 6 Distribution of Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways |
Asish Kumar Parida, A. B. D. (2004). Salt tolerance and salinity effects on plants: a review. Ecotoxicology and Environmental Safety, 60(3), 324-349. doi: 10.1016/j.ecoenv.2004.06.010
http://dx.doi.org/10.1016/j.ecoenv.2004.06.010
Changyou Li, Bin Fang, Chunwu Yang, Decheng Shi,Wang, D. (2009). Effects of Various Salt–Alkaline Mixed Stresses on the State of Mineral Elements in Nutrient Solutions and the Growth of Alkali Resistant Halophyte Chloris Virgata. Journal of Plant Nutrition, 32(7), 1137-1147. doi: 10.1080/01904160902943163
http://dx.doi.org/10.1080/01904160902943163
Chao DongHui, 2012, CThe studies of DNA methylation occuured in alkali-resisitant halophyte Chloris virgata and glycophyte cotton under kinds of salt and alkali stresses, Dissertation for Ph.D., Northeast Normal University
Cheng-Ying Shi, Hua Yang, Chao-Ling Wei, Oliver Yu, Zheng-Zhu Zhang, Chang-Jun Jiang, et al. (2011). Deep sequencing of the Camellia sinensis transcriptome revealed candidate genes for major metabolic pathways of tea-specific compounds. BMC Genomics, 12:131. doi: 10.1186/1471-2164-12-131
http://dx.doi.org/10.1186/1471-2164-12-131
Conesa A, Götz S, García-Gómez JM, Terol J, T. M.,M., R. (2005). Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics, 21(18), 3674-3676. doi: 10.1093/bioinformatics/bti610
http://dx.doi.org/10.1093/bioinformatics/bti610
Consortium., G. O. (2008). The Gene Ontology project in 2008. Nucleic Acids Research, 36, 440-444. doi: 10.1093/nar/gkm883
http://dx.doi.org/10.1093/nar/gkm883
Dai Gaoxing, Peng Keqin,Canhui, P. (2003). The Effects of Calcium on Salt -tolerance in Plant. Chinese Agricultural Science Bulletin, 19(3), 97-101.
Dong Lili, Gong Chengxia,Weiguo, S. (2015). On Restoration and Improvement of Saline-alkali Soil. TIANJIN SCIENCE & TECHNOLOGY, 42(8), 68-72.
Du J, Yuan Z, Ma Z, Song J, Xie X,Y, C. (2014). KEGG-PATH: Kyoto encyclopedia of genes and genomes-based pathway analysis using a path analysis model. Mol Biosyst., 10(9), 2441-2447. doi: 10.1039/c4mb00287c.
http://dx.doi.org/10.1039/C4MB00287C
Dutkowski J,J, T. (2007). Identification of functional modules from conserved ancestral protein-protein interactions. Bioinformatics, 23(13), 149-158. doi: 10.1093/bioinformatics/btm194
http://dx.doi.org/10.1093/bioinformatics/btm194
Harris MA, Clark J, Ireland A, Lomax J, Ashburner M, Foulger R, et al. (2004). The Gene Ontology (GO) database and informatics resource. Nucleic Acids Research(32), 258-261. doi: 10.1093/nar/gkh036
http://dx.doi.org/10.1093/nar/gkh036
Jin, H. K., H.R.Plaha, P. Liu, S.K. Park, J.Y. Piao, Y.Z. Yang, et al. (2008). Expression profiling of the genes induced by Na2CO3 and NaCl stresses in leaves and roots of Leymus chinensis. Plant Science, 175(6), 784-792.
http://dx.doi.org/10.1016/j.plantsci.2008.07.016
Lin CM,W, F. (2012). Microarray and synchronization of neuronal differentiation with pathway changes in the Kyoto Encyclopedia of Genes and Genomes (KEGG) databank in nerve growth factor-treated PC12 cells. Curr Neurovasc Res, 9(3), 222-229.
http://dx.doi.org/10.2174/156720212801618983 PMid:22697417
Moriya Y, Itoh M, Okuda S, Yoshizawa AC,M, K. (2007). KAAS: an automatic genome annotation and pathway reconstruction server. Nucleic Acids Research(35), 182-185. doi: 10.1093/nar/gkm321
http://dx.doi.org/10.1093/nar/gkm321
Nishiuchi S, Liu S,T, T. (2007). Isolation and characterization of a metallothionein-1 protein in Chloris virgata Swartz that enhances stress tolerances to oxidative, salinity and carbonate stress in Saccharomyces cerevisiae. Biotechnol Lett, 29. doi: 10.1007/s10529-007-9396-4
http://dx.doi.org/10.1007/s10529-007-9396-4
Qingmei Guan, Jianmin Wu, Xiule Yue, Yanyan Zhang,Zhu, J. (2013). A nuclear calcium-sensing pathway is critical for gene regulation and salt stress tolerance in Arabidopsis. PLOS Genetics, 9(8), 1-16. doi: 10.1371/journal.pgen.1003755
http://dx.doi.org/10.1371/journal.pgen.1003755
Shunsaku Nishiuchi, Kazumasa Fujihara, Shenkui Liu,Takano, T. (2010). Analysis of expressed sequence tags from a NaHCO3-treated alkali-tolerant plant, Chloris virgata. Plant Physiology and Biochemistry, 48, 247-255.
http://dx.doi.org/10.1016/j.plaphy.2010.01.024
Wang Jiali, Huang Xianjin, Zhong Taiyang,Zhigang, C. (2011). Review on Sustainable Utilization of Salt-affected Land. ACTA GEOGRAPHICA SINICA, 66(5), 673-684.
Xi Zhang, Junbo Zhen,Li, Z. (2011). Expression Profile of Early Responsive Genes Under Salt Stress in Upland Cotton (Gossypium hirsutum L.). Plant Molecular Biology Reporter, 29(3), 626-637.
http://dx.doi.org/10.1007/s11105-010-0269-y
Yiou Pan, Chen Yang, Xiwu Gao, Tianfei Peng, Rui Bic, Jinghui Xi, et al. (2015). Spirotetramat resistance adaption analysis of Aphis gossypii Glover by transcriptomic survey. Pestic Biochem Physiol, 124, 73-80. doi: 10.1016/j.pestbp.2015.04.007
http://dx.doi.org/10.1016/j.pestbp.2015.04.007
Zhu, J.-K. (2001). Plant salt tolerance. Trends in Plant Science, 6(2), 66-71. doi: 10.1016/S1360-1385(00)01838-0
. PDF(639KB)
. FPDF(win)
. HTML
. Online fPDF
Associated material
. Readers' comments
Other articles by authors
. Yajun Sun
. Xiaoxue Ye
. Panpan Liu
. Lee ImShik
Related articles
. Chloris virgate
. cDNA library
. Gene ontology annotation
. Stresses, COG
. Pathways
Tools
. Email to a friend
. Post a comment


